Moving into the fall semester of my Sophomore year at Virginia Tech, I was a little starved of side projects I could do, simply due to the amount of schoolwork I had to finish each week. However, luckily I was able to make time for a couple projects I could focus on in the side. One of them being is data analytics competition, of which was sponsored by the CMDA club here at VT and GDMS. I participated in the beginners competition in a group of 3, the other members being good friends of mine Eric Fu and Kali Liang. In the competition, we were given a large data set (.csv file) involving health issues across various cities throughout the United States (500 entries). Some examples of the categories, just to get a feel of the type of data we received, included ailments such as asthma, obesity, heart disease, kidney disease, etc. We were also given general data involving disease prevention (i.e. access to health care, going to regular checkups, etc.) and unhealthy behaviors (i.e. binge drinking, drug use, lack of leisure time and sleep, etc.).
I believe that my group’s approach to the data was a rather unique one, given our different backgrounds. Eric is a BIT major, Kali is a CMDA (Computational Modeling and Data Analytics) and Statistics major, and I am, of course, a Computer Science major. As a result of our different backgrounds, we each had different ideas as to how to approach the information to find correlations between the categories. For instance, my approach was to code basic programs in Python using numpy and matplotlib to generate scatterplots and other useful graphs, while Kali was comfortable using R to find more statistics-heavy information (like GLM/LS Means) and Eric was comfortable working directly in the given Excel file to look for correlations.
First, we decided as a group to look for interesting differences in the data by splitting the it up into regions. We explored a number of splits, and we ended up using data from a regional split (NE, S, MW, W), a coastal split (west/east coast, with everything else being lumped into a “main land” group), and a split based on population size to explore the uniqueness of big cities (we defined a big city as any city above 2 standard deviations above the mean population for the entire data set). We also explored a split based on State, but found the data to be too volatile, and with 51 groups (incl. Washington DC), there were still too many points to investigate. My python scripts came into good use here, as I was able to make a quick program that found and graphed the scatterplots of each region for any given category, along with a line that connected the means for each sub group together (with and without outliers) for visual investigation. I also coded a script that plotted scatter plots for any 2 given categories to see if they were related.
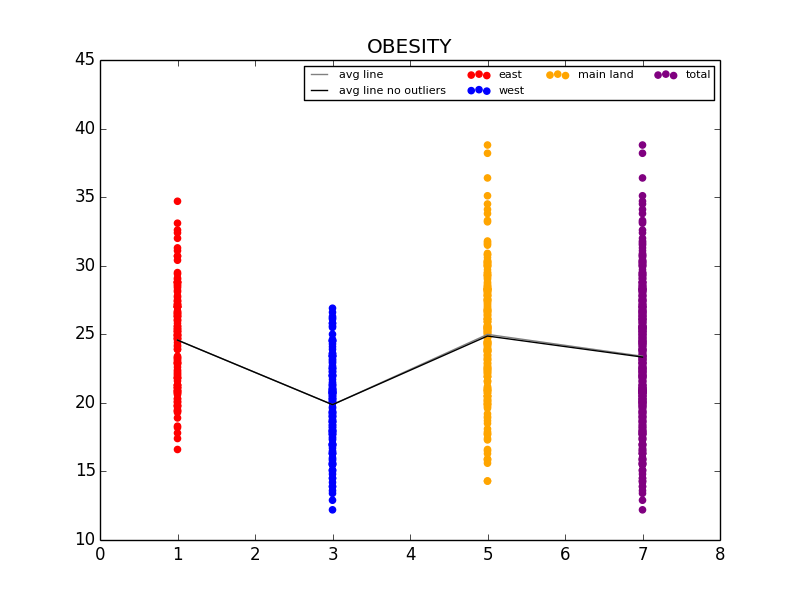
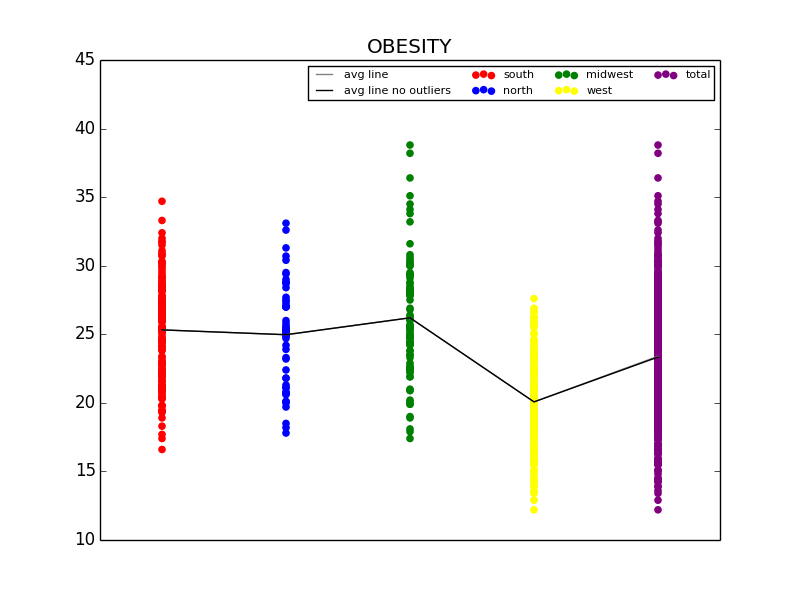
The first graph details the difference in Obesity in the east vs. west coast, while the second illustrates the difference in Obesity in the US Regions.
From this initial stab at the data, we found what we believed to be significant differences between the general health in the east coast and the west coast (meaning the west coast was significantly lower than the east in almost all categories). Also worth noting, we also determined that health in the west region was generally better than the other regions. From this lead, we explored the differences in the means to attempt to understand why the west is generally healthier. We observed that both the west coast and the west region have significantly lower Obesity rates when compared to other regions, and decided to explore that as a possibility (see the mean comparison charts above). We confirmed our assumption by looking at the general trends for obesity and various other ailments and observed that, in general, there is a strong link between obesity and other serious health problems.
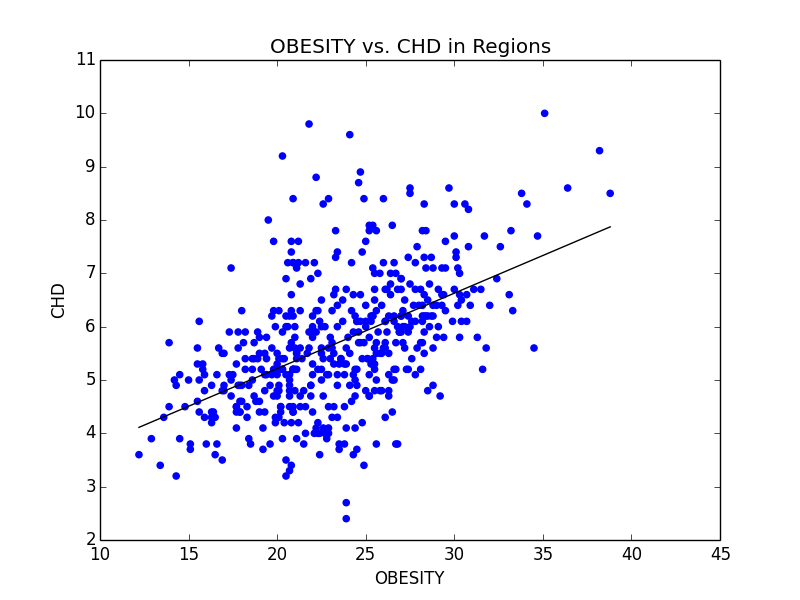
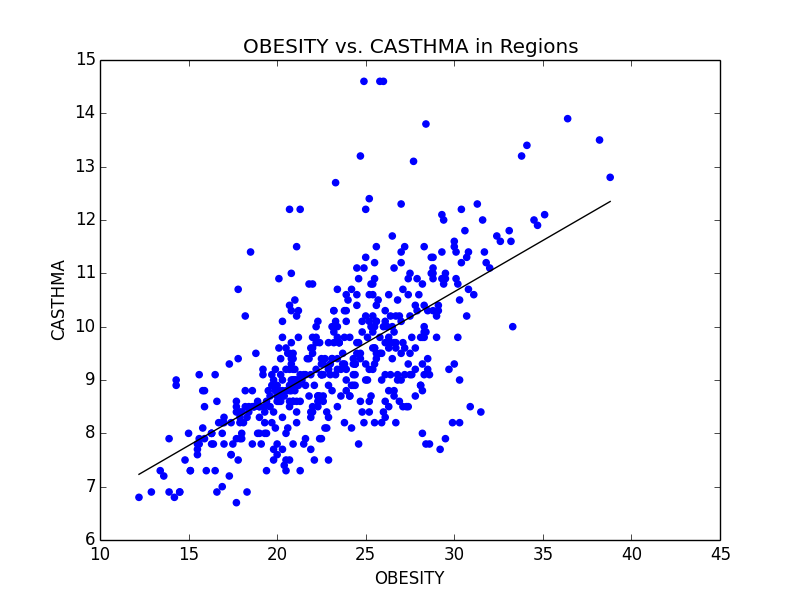
Some of the scatterplots we used to illustrate the link between Obesity and other serious illnesses like heart disease (CHD) and asthma (CASTHMA).
In order to further provide evidence for our assumptions, Kali used regression analysis, specifically generalized linear models with least square means to further look for significance between these groups. Using these tests was helpful because it not only aided at eliminating some of the confounding variables that may be lurking within the data, but made it extremely clear as to whether the data was significant or not through looking at the p-value numerically and with a visual aide. Essentially, when graphed, intervals of the mean are plotted for each region (each interval determined using a 95% CI). If the regions don’t have any overlapping points, then the difference between the two can be considered significant and is definitely worth looking into. As you can see below, we were able to confirm our assumption that the difference between obesity rates in the east and west is significant.
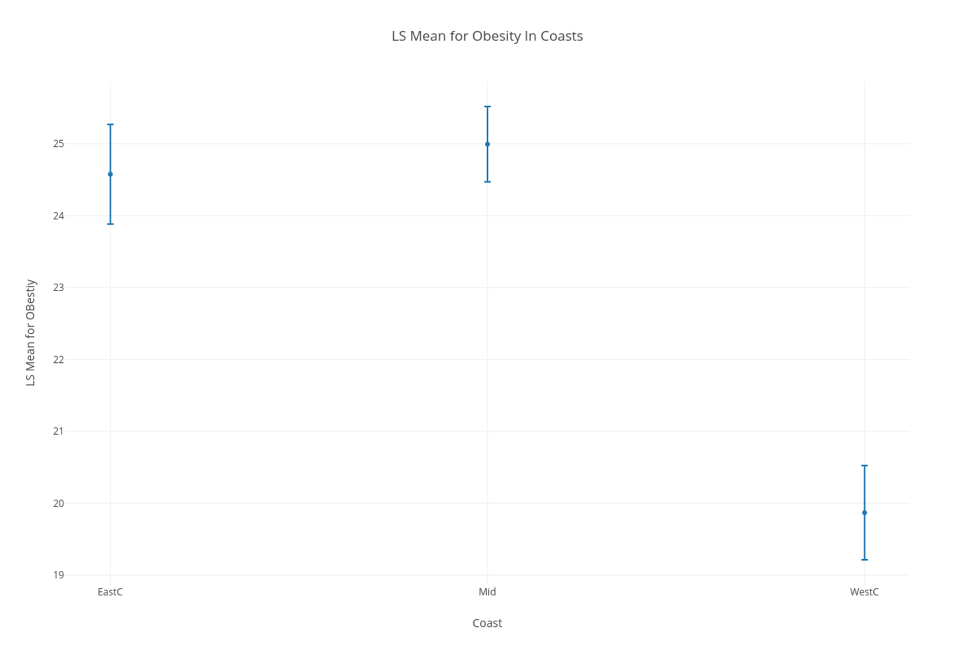
LSM Chart confirming our assumption that the difference in Obesity rates in the east and west coasts is significant
However, we were not satisfied simply stating that we recommend that Obesity rates be lowered, so we looked into possible variables that could affect Obesity that weren’t already included in our data set. Ideas for this included finding density of fast food restaurants within each city, and measuring the nutritional differences between each city. Unfortunately, we were unable to find any reliable data on density of fast food restaurants, but we were able to pull data from the CDC website pertaining to amount of people that ate less than one fruit and one vegetable per state in the US. Using this data, we first confirmed the link between not eating nutritional food and obesity, then went on to make our recommendation. Eric’s background in Business came in handy, as we gave various recommendations that made sense from an economics point of view (i.e. decrease the tax or cost of fruits and vegetables in obese regions, subsidize farmers to increase fruit and vegetable output, or push advertisements encouraging people to eat healthier).
As a slight aside to our main focus on Obesity across the US, we also found that Stress (we defined stress as a lack of leisure time (LPA) and a lack of sleep) was high in big cities. We noticed that not only were kidney problems slightly higher in bigger cities, but so were other ailments such as strokes and diabetes. We were able to link the connection between stress and diabetes with kidney problems (seen below), and further connect kidney problems with strokes. Unfortunately, due to the time constraints for the competition, we were not able to explore these connections much deeper. However, given more time, we definitely would have done so.
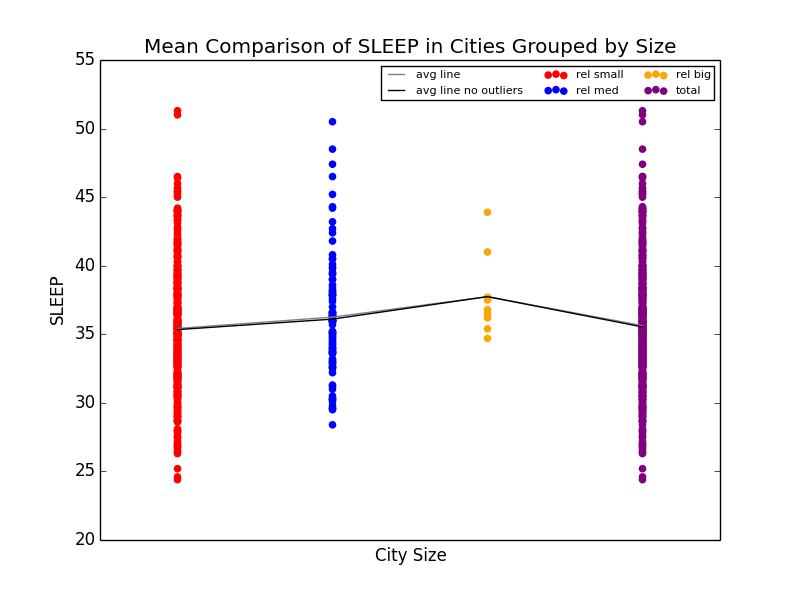
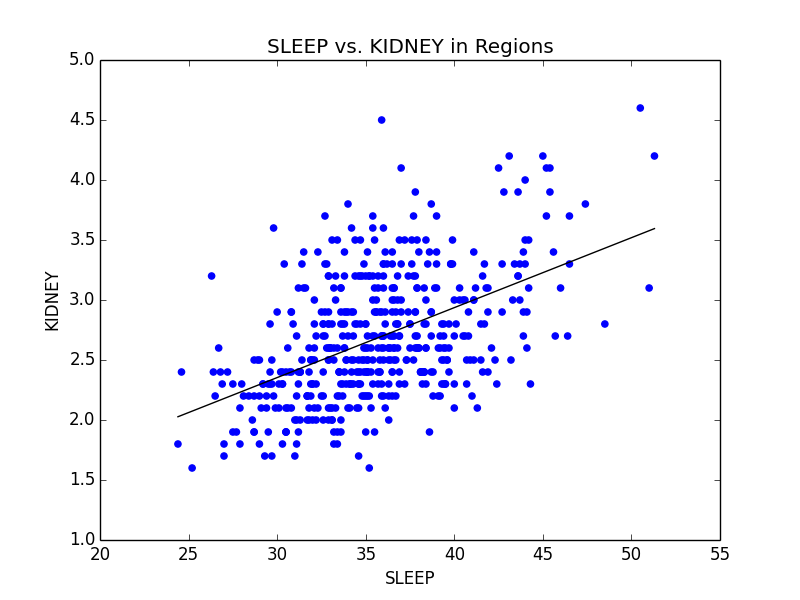
Mean comparison showing increase in lack of sleep in big cities and scatterplot showing the relationship between sleep deprivation and kidney problems.
This competition was extremely fun to participate in. Unfortunately, we did not place in the top 3 teams for our section, although we did get an honorable mention. I want to thank GDMS and the CMDA club at VT for making this a reality. I’ve always been interested in data analytics due to my background in neural networks, and it was incredibly fun to work on something data analytics related even though I haven’t been able to take any statistics courses as of yet.
If you’d like to see any more plots, or would like to see the scripts I made for the purpose of this competition, feel free to email me at [email protected].







Recent Comments